Google Summer of Code 2022, CuPy, Talk
Khushi Agrawal
@khushi-411
@khushi__411
This talk on Github: khushi-411/gsoc-cupy-talk
About CuPy
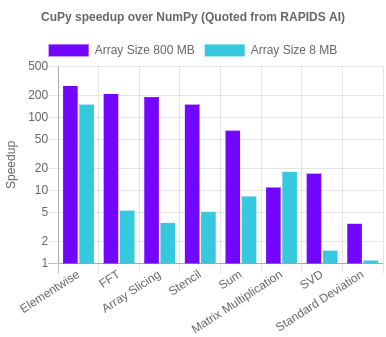
NumPy/SciPy-compatible Array Library for GPU-accelerated Computing with Python. CuPy speeds up some operations more than 100X. cupy.dev
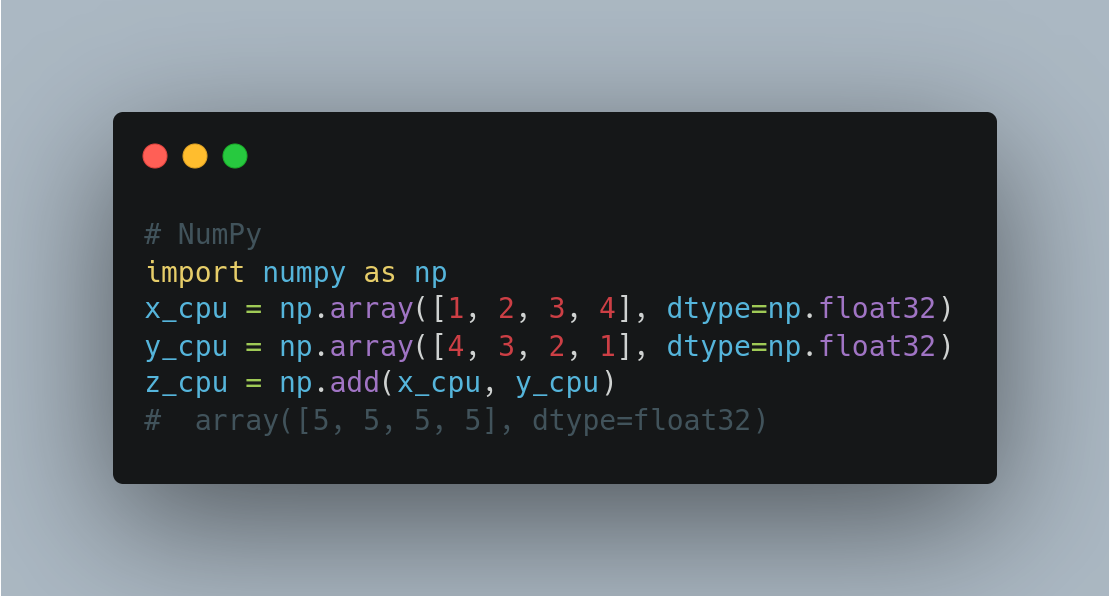
NumPy (GPU not supported)
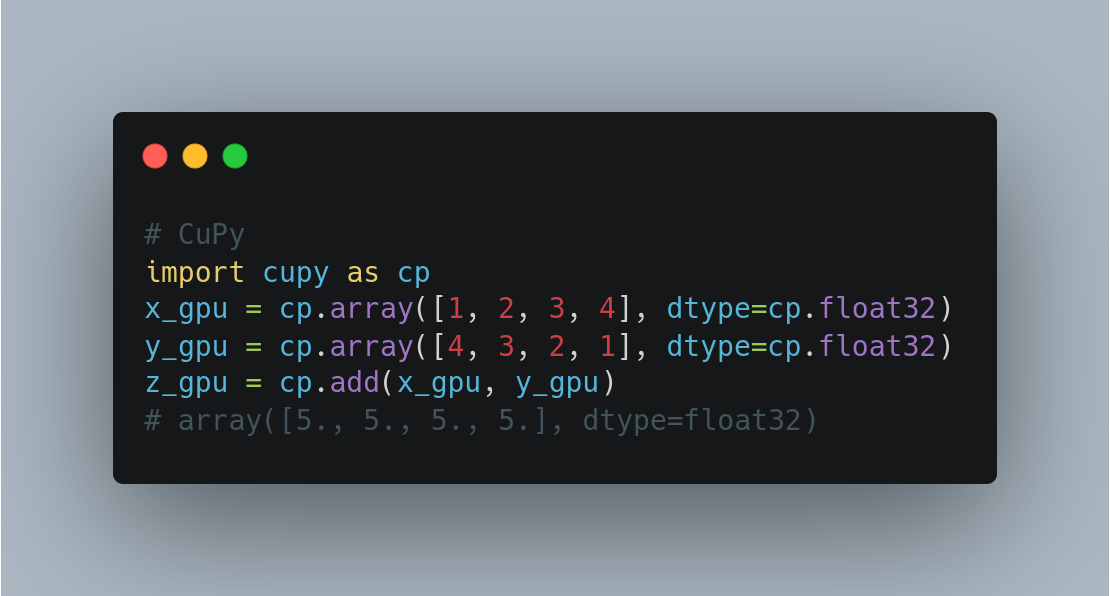
CuPy (GPU supported)
Projects using CuPy: https://github.com/cupy/cupy/wiki/Projects-using-CuPy
The Project
Project 1: CuPy coverage of NumPy/SciPy functions
Mentor: Masayuki Takagi
Expected Outcomes
Optimized GPU implementation of NumPy/SciPy APIs
Documentation and tests
Performance Benchmarking (use NVIDIA Nsight System)
Functions I Proposed & Accomplished
Interpolate
BarycentricInterpolator
KroghInterpolator
Special
log_ndtr
expn
softmax
log_softmax
Stats
boxcox_llf
zmap
zscore
Other Goals Accomplished (apart from proposed functions)
Add complex number support for nanvar & nanstd
logsumexp
barycentric_interpolate
krogh_interpolate
WIP PR
support multiple output args in ReductionKernel
Pre-GSoC Time
Enhanced CuPy’s coverage of NumPy functions. My contribution’s:
- #6571 Add cupyx.scipy.special.{i0e, i1e} (Thanks @takagi!)
- #6433 Add cupy.setdiff1d (Thanks @asi1024!, Thanks @kmaehashi!, Thanks @emcastillo!)
- #6402 Add cupy.intersect1d (Thanks @asi1024!)
- #6357 Add cupy.union1d (Thanks @takagi!)
- #6316 Add triu_indices and triu_indices_from (Thanks @takagi!)
- #6305 Add triu_indices and triu_indices_from (Thanks @takagi!)
- #6294 Add doc issue template (Thanks @kmaehashi!)
- #6282 Add cupy.fabs (Thanks @toslunar!)
- #6280 Add cupy.ediff1d (Thanks @takagi!)
- #6275 Add asarray_chkfinite (Thanks @kmaehashi!, Thanks @leofang!, Thanks @toslunar!)
- #6254 Add array_equiv (Thanks @kmaehashi!)
- #6089 Add cupy.isneginf and cupy.isposinf (Thanks @kmaehashi!)
- #6085 Add cupy.asfarray (Thanks @kmaehashi!)
Interpolate
Introduced interpolate module in CuPy.
Before:
import cupy
from cupyx.scipy import interpolate
# throws error
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
ImportError: cannot import name 'interpolate' from 'cupy' (/home/khushi/Documents/cupy/cupy/__init__.py)
Now:
import cupy
from cupyx.scipy import interpolate
# works well
BarycentricInterpolator & barycentric_interpolate
Paper: https://people.maths.ox.ac.uk/trefethen/barycentric.pdf
Added
_Interpolator1Dclass: deals with common features for all interpolation functions (implemented in CPU, due to a smaller number of points). It supports the following methods:-
__call__: use to call the next points -
_prepare_x: change into a 1-D array -
_finish_y: reshape to the original shape -
_reshape_yi: reshape the updated yi values to a 1-D array -
_set_yi: if y values are not provided, this method is used to create a y-coordinate -
_set_dtype: sets the dtype of the newly created yi point -
_evaluate: evaluates the polynomial, but for reasons of numerical stability, currently, it is not implemented.
-
Added
BarycentricInterpolatorclass: constructs polynomial. It supports the following methods:-
set_yi: update the next y coordinate, implemented in CPU due to smaller number of data points -
add_xi: add the next x value to form a polynomial, implemented in CPU due to smaller number as mentioned in the paper -
__call__: calls the _Interpolator1D class to evaluate all the details of the polynomial at point x -
_evaluate: evaluate the polynomial
-
Added
barycentric_interpolatewrapper
KroghInterpolator & krogh_interpolate
Added
_Interpolator1DWithDerivativesclass: calculates derivatives. Its’ parent class is_Interpolator1D. It supports the following methods:-
derivatives: evaluates many derivatives at point x -
derivative: evaluate a derivative at point x
-
Added
KroghInterpolatorclass: constructs polynomial and calculate derivatives. It supports the following methods:-
_evaluate: evaluates polynomial -
_evaluate_derivatives: evaluates the derivatives of the polynomial
-
Added
krogh_interpolatewrapper
Special
Enhanced the coverage of special functions in CuPy.
- APIs covered:
-
log_ndtrPR #6776- For more details: https://khushi-411.github.io/GSoC-week1/
-
log_softmaxPR #6823 -
logsumexpPR #6773-
support multiple output args in reduction kernelPR #6813
-
-
expnPR #6790- GitHub repository for more details: https://github.com/khushi-411/expn
-
softmaxPR #6890
-
- Documentations and tests
- Performance Benchmark
Universal Functions
- Implemented in
log_ndtr&expn(both numerically stable) - Wraps into C++ snippet for speedy implementation.
# universal function
log_ndtr = _core.create_ufunc(
'cupyx_scipy_special_log_ndtr',
(('f->f', 'out0 = log_ndtrf(in0)'), 'd->d'),
'out0 = log_ndtr(in0)',
preamble=log_ndtr_definition, # C++ function call
doc="""
....
"""
)
Custom Kernels (ReductionKernel)
- Similar to
map&reduceoperations in python. - Wraps into C++ snippet for speedy implementation.
# function call
def(...):
...
_log_softmax_kernel(tmp, axis=axis, keepdims=True)
return ...
# ReductionKernel
_log_softmax_kernel = cp._core.ReductionKernel(
'T x1',
'T y',
'exp(x1)',
'a + b',
'y = log(a)',
'0',
name='log_softmax'
)
Kernel Fusion (Experimental)
- Compiles into a single kernel instead of series of kernels
- Experimental implementation to fuse
softmaxfunction - Drawback here: does not generate competitive codes, therefore, switched to the normal implementation
def make_expander(shape, axis):
axis = internal._normalize_axis_indices(axis, len(shape))
expander = []
for i, s in enumerate(x.shape):
if i in axis:
expander.append(None)
else:
expander.append(slice(None))
return tuple(expander)
TODO: Add shape method in cupy.fusion, to use make_expander inside fusion kernel
@_util.memoize(for_each_device=True)
def _softmax_fuse(shape, axis):
expander = make_expander(shape, axis)
@_core.fusion.fuse()
def softmax_fuse(x):
x_max = cupy.amax(x, axis=axis)
exp_x_shifted = cupy.exp(x - x_max[expander])
return exp_x_shifted / cupy.sum(exp_x_shifted, axis=axis)[expander]
return softmax_fuse
def softmax(x, axis=None):
fused_softmax = _softmax_fuse(shape=x.shape, axis=axis)
return fused_softmax(x)
Stats
Enhanced the coverage of statistical functions in CuPy.
boxcox_llfPR #6849zmap & zscorePR #6855- Complex number support in
nanvarandnanstdPR #6869- For more details: https://khushi-411.github.io/GSoC-week6/
- Complex number support in
Acknowledgement
Thanks in millions to Masayuki Takagi for guiding me throughout the GSoC timeline and even before joining GSoC. I really appreciate you for being an incredibly supportive mentor. I am pleased and glad to get in touch with you!
Thank you!
